
email:
roberto_kolter@hms.harvard.edu
Biosketch (PDF)
Google Scholar
From 1983, when I joined Harvard Medical School as an Assistant Professor until 2018, when I retired and closed the lab, I was deeply involved in research and education. More than 130 individuals trained in my laboratory during those 35 years and most of those trainees followed careers in science in both academic and industry settings. Research in the Kolter Lab always gravitated around the study of microbes. We explored a large number of different subjects ranging from basic bacterial physiology to bioactive compound discovery.
 Now, as Professor Emeritus I continue my involvement in science through teaching, writing and blogging (at Small Things Considered) and communicating microbial sciences to the public, including through exhibitions in museums of natural history and invited lectures. Our photographic exhibition World in a Drop, produced jointly with Scott Chimileski and first shown at the Harvard Museum of Natural History in 2017, continues to travel to numerous locations around the world. Our exhibition Microbial Life is the major special exhibition at the Harvard Museum of Natural History for two years, open from February 2018 to March 2020. Our book Life at the Edge of Sight (co-authored with Scott Chimileski) was released by Harvard University Press September 2017.
Now, as Professor Emeritus I continue my involvement in science through teaching, writing and blogging (at Small Things Considered) and communicating microbial sciences to the public, including through exhibitions in museums of natural history and invited lectures. Our photographic exhibition World in a Drop, produced jointly with Scott Chimileski and first shown at the Harvard Museum of Natural History in 2017, continues to travel to numerous locations around the world. Our exhibition Microbial Life is the major special exhibition at the Harvard Museum of Natural History for two years, open from February 2018 to March 2020. Our book Life at the Edge of Sight (co-authored with Scott Chimileski) was released by Harvard University Press September 2017.
Summary of Scientific Contributions
View all publications at PubMed or Google Scholar
As a graduate student (1975-1979) I studied the control of plasmid DNA replication. The results of my work provided some of the earliest direct physical evidence for the "replicon hypothesis" that had been put forth in 1962 by Jacob, Brenner and Cuzin. In my work, I was able to separate (and thus define) an origin of DNA replication from a gene encoding a initiator protein that bound and activated the origin. Aside from the basic knowledge provided, this separated replicon served as the foundation for many of the "suicide cloning vectors" still in wide use today. When I started my laboratory at HMS in 1983, the genetic bases for antibiotic biosynthesis and export was virtually unexplored. I developed a program to identify and characterize the genes involved in the production of small peptide antibiotics produced by Escherichia coli. The power of the genetic tools available in this organism and the application of the nascent technologies of rapid cloning and sequencing allowed us to lead the field in the area of peptide antibiotic biosynthesis. We discovered several novel post-translational modifications and were among the first to characterize the so-called "ABC Exporters", now known to be among the most widespread membrane proteins involved in both import and export processes. Prior to the mid-1980s, the majority of studies of bacterial physiology were done on cultures growing in exponential phase or in chemostats. Realizing that in their natural settings bacteria seldom encounter such conditions, we began to study bacteria in stationary phase. This way we discovered regulatory systems that operated only in non-growing cells whose net effect was to render the cells more resistant to diverse stresses. These results had such an impact that a new Gordon Conference was formed to discuss "Microbial Stress Responses". When the conference celebrated its 20th year, I was invited to present the meeting’s opening Neidhardt lecture. Perhaps the most suprising finding we made was the discovery that stationary phase cultures were remarkably dynamic, with fitter mutants taking over in a matter of a few days. This was one of the early examples of experimental evolution. By the mid-1990s my laboratory opened another field of study in bacterial physiology by applying genetic approaches to understand bacterial biofilms. Prior to our work, very few bacterial geneticists and molecular biologists were studying these surface-associated microbial communities. The vast majority of the studies involving biofilms were carried out by engineers interested in flow dynamics and how these were influenced by fouling. Our recognition that most microbes in natural settings are indeed surface-associated led us to begin to characterize the process of biofilm formation by performing mutant screens in many species, looking for biofilm-defective mutants. We published two seminal papers in 1998 and an influential review in 2000, followed by dozens more over then next two decades. During that time, ours was considered one of the key biofilm research laboratories. Since the early days of microbiology the discipline has been greatly influenced by the studies of pure cultures. Yet, it has always been recognized that microbes seldom exist as pure cultures in natural settings. At the turn of the century we began pioneering the use of co-cultures to understand how microbes alter their physiologies as a consequence of the presence of other species. The study of the emerging properties of multi-species communities went on to occupy the attention of many members of my lab and our results greatly influenced how others characterized microbial interactions.1. Regulation of DNA Replication

2. Peptide Antibiotic Biosynthesis

3. Physiology and Evolution During Stationary Phase
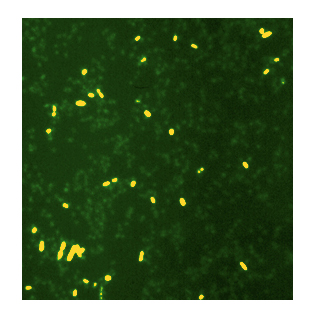
4. Bacterial Biofilms
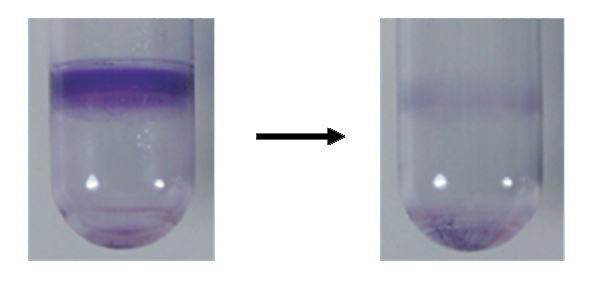
5. Microbial Interspecies Interactions





